
How are protective SNPs identified? Can they be identified through the GWAS?
February 18, 2022

- Related Topics:
- Genetic variation,
- Population genetics,
- Bioinformatics
A curious adult from Georgia asks:
“After reading your article ”How GWAS works" I have a related question. How are protective SNPs identified? Can they be identified through the GWAS?”
Yes, protective SNPs can also be identified through GWAS! In fact, for each SNP, a protective allele is almost always identified along with a risk allele. Let’s see why.
G-WhAt?
First, let’s do a quick recap of Genome-Wide Association Studies (GWAS).
One way to study disease genetics is to compare people who have a specific disease to people who do not have that disease. Researchers sequence each person’s DNA in order to identify any significant differences between the two groups.
Since these studies require a LOT of participants, sequencing the entire genome would be too expensive. Instead, most of the time, researchers focus on about a million different places in your DNA called single-nucleotide polymorphisms (SNPs).
As their name suggests, they’re just a single nucleotide (base). That is, a single A, C, T, or G. They’re also polymorphic in the population. This just means that some people have one allele (perhaps A) while other people have a different allele (like G).
GWAS studies are looking for SNPs that differ in frequency between the disease and non-disease groups. Researchers performing a GWAS usually don’t start out with any idea of where these differences might be. So they search SNPs across the whole genome — thus, “Genome-Wide”.
The differences that they discover are called “Associations” because particular SNPs are associated with having the disease or not. Each association could be “risk” if the SNP increases your chances of getting the disease, or “protective” if it decreases your chances. Now we see how Genome-Wide Association Studies got their name!
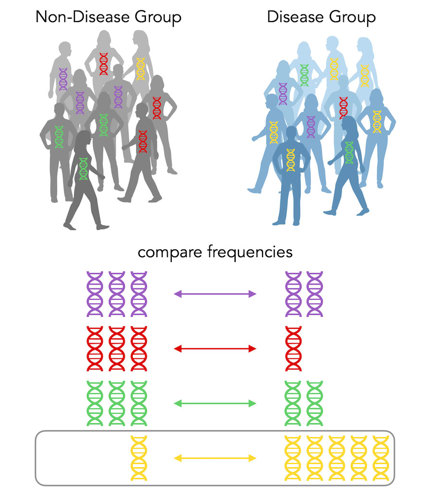
Shades of gray
In the above case, there is a lot more yellow DNA in the disease group, so we say that yellow is a risk allele for the disease. It’s that easy!
Wait, no it’s not.
One person has the yellow DNA but is in the non-disease group. Were they put there by mistake? Well, no. GWAS associations are not black-and-white, yes-or-no. In this case, the yellow DNA increases the chances you’ll get the disease, but it still isn’t guaranteed.
The article you mentioned in your question talks a lot about the odds ratio, which is a measure of how risky that risk factor actually is. A ratio above 1 means the allele is a risk factor, while a ratio below 1 means the allele is protective.
Let’s calculate the odds ratio. There are 5 people with the yellow allele in the disease group and only 1 person with the yellow allele in the non-disease group. I’ve summarized this in the table below.
|
|
Disease |
Non-Disease |
|
Yellow |
5 |
1 |
|
Not Yellow |
5 |
9 |
The odds ratio is a ratio: On the top, we have the odds of getting the disease if you have the yellow allele, and on the bottom, we have the odds of not getting the disease if you have the yellow allele.
So, we have (5/5) ÷ (1/9) = 9. So in this case, the odds ratio for the yellow allele is 9. That’s way more than 1! So the yellow allele is super duper risky for the disease.
But what about protective alleles? Let’s look at another scenario:
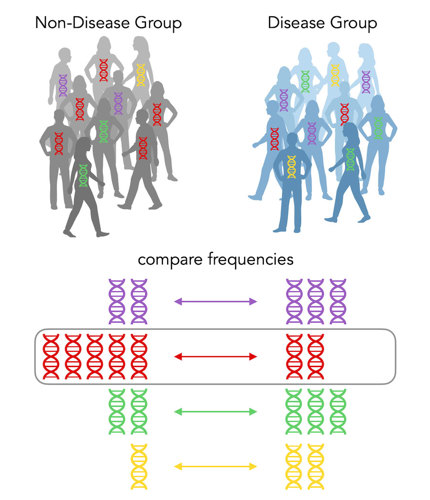
In this case, there isn’t really a significant increase for any color in the disease group. BUT, there’s a significant increase in red DNA in the non-disease group. Its odds ratio is (2/8) ÷ (5/5) = ¼. It’s less than 1! So the red allele is indeed a protective allele. It protects you from getting the disease!
One in a Million
When scientists perform a GWAS, they’re doing the same thing as in the diagrams above, but with each of the million SNPs one-at-a-time.
Usually, there’s just two alternatives for each SNP (in my example, A or G). Rarely, there could be three or even all four nucleotide options at that SNP. But most of the time, there’s just two.
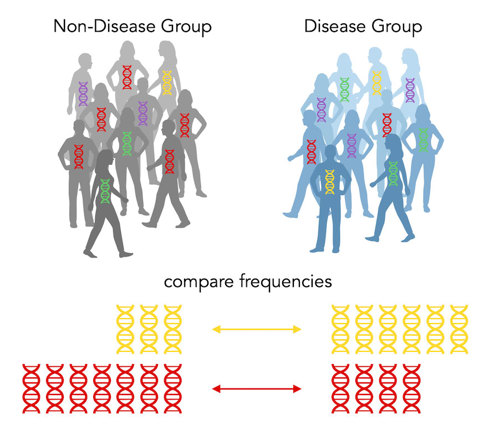
Above, we see that the disease group has more of the yellow allele, while the non-disease group has more of the red allele. So yellow is a risk allele and red is a protective allele. The yellow has an odds of 3 and the red has an odds ratio of ⅓. (Note: when there’s only two options, the odds ratios will be reciprocals.)
Because there are only two alternatives (red or yellow), every risk allele by definition has to have a protective allele counterpart! Amazing!
Wait, if every SNP has BOTH a protective AND a risk allele, why do we report an entire SNP as a risk SNP? That’s like reporting only one side of a story.
When there’s only two options, they’re only risky or protective relative to one another. So we have to pick what the baseline is going to be.
Every SNP is Both???
Unfortunately, the baseline is rather arbitrary. It’s based on the human reference genome — which was sequenced way back in 2001 from only a handful of humans.
Let’s say that the red allele is present in the human reference genome. So we’d refer to red as the “reference” allele. Which makes yellow the “alternative” allele. Note that this doesn’t necessarily have anything to do with the frequency of each of these alleles. Yellow could be present in 99% of people, yet could still be called the “alternative” allele if it wasn’t in the reference genome.
For GWAS, we usually report what the alternative allele is up to. In this case, the alternative yellow allele is a risk allele, so we’d call this risk SNP. If yellow was the reference allele, then we’d call this a protective SNP.
I know, it’s confusing. Articles tend to throw around the terms “risk SNP” or “protective SNP” willy-nilly. It’s more precise to say that a SNP is associated with a disease, but a particular allele is either risky or protective.
Many SNPs, One Disease
Most traits and diseases are complex — that is, they are not caused by a single gene, but rather a lot of genes and environmental factors all working together.
It’s common for a GWAS study to report dozens or even hundreds of SNPs associated with a particular disease! Because there are so many SNPs that affect a complex disease, you’ll likely have the protective allele at some of these SNPs and the risk allele at others.
As an example, let’s consider Type 2 Diabetes. The latest GWAS in European individuals identified 1227 associated SNPs. Of these, 583 SNPs were risk and 644 were protective.1,2
But even with over a thousand SNPs associated with Type 2 Diabetes, genetics only accounts for 25-75% of the risk for the disease. Why so low? Well, age, Body Mass Index (BMI), fast-food intake, and exercise frequency also play a role.2
A person with a lot of risk alleles who exercises frequently and has a healthy BMI may never develop Type 2 Diabetes, while a person who has a lot of protective alleles but poor lifestyle choices can still develop the disease.
Thus, it’s easy to over-interpret your health results from 23andMe. It’s really cool to see if you carry risk or protective SNPs for certain diseases, but context is key!

Author: Alyssa Lyn Fortier
When this answer was published in 2021, Alyssa Lyn was a Ph.D. candidate in the Stanford Department of Biology, studying the evolution of immunity-related genes in Jonathan Pritchard’s laboratory. She wrote this answer while participating in the Stanford at The Tech program.
 Skip Navigation
Skip Navigation
