
How does Next Generation Sequencing work?
July 2, 2019

- Related Topics:
- Biotechnology,
- DNA sequencing,
- Genomics
An undergraduate student from NY asks:
"How does Next Generation Sequencing work? Technically, how is NGS better than Sanger Sequencing? I am a little bit confused on how the dNTPs play a role in identifying base pairs."
The ability to read the sequence of DNA code has revolutionized biology in recent decades. It has led to massive insights in the understanding of biology and disease.
As scientists have improved on sequencing techniques, it has become even cheaper and easier to read DNA sequences. With falling costs has come an explosion in demand: DNA sequencing is now a routine tool used in laboratories around the world.
Let's see how Sanger sequencing compares to more recent methods (Next Generation Sequencing, NGS).
What is Sanger sequencing?
Sanger sequencing was first developed by Frederick Sanger in the 1970s. It was the first sequencing method to be commercialized, and it is still widely used today.
Most sequencing techniques, including Sanger methods, are based on the natural process used by a cell to copy its DNA.
When a cell copies its DNA, it uses a special enzyme called polymerase. First, the cell unwinds its DNA into two separate single strands. Polymerase binds to this single stranded DNA and fills in one base at a time. As it adds the nucleotides (dNTPs; A, T, G, or C), it fills in the single-stranded DNA to be a full piece of double-stranded DNA. (Read more about DNA replication here.)
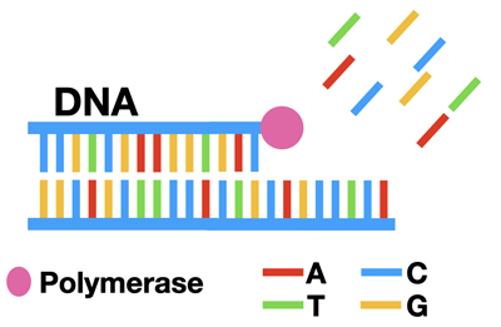
Scientists figured out that you can use this process to copy any piece of DNA in a test tube. Through Polymerase Chain Reaction (PCR) it's possible to make lots and lots of copies of any DNA sequence! (Read more about PCR here.)
Sanger Sequencing is based on PCR. If you can read each DNA letter as it's added, you can figure out what the sequence is.
To do this, we'll need to control the DNA replication process. Ideally, something with a label that can easily show which base is added.
Sanger sequencing uses special nucleotide bases called dideoxynucleotides (ddNTPs). These mostly look like regular DNA bases (dNTPs), with one small chemical change.
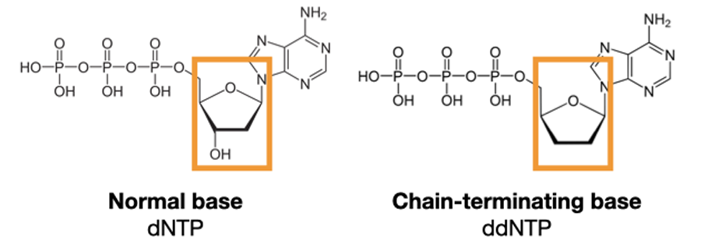
This small change makes a big difference. It removes the attachment place for new DNA bases! Polymerase can add these special bases to a growing strand of DNA. But once it's added, the chain is stopped. That's why they're often called chain-terminating nucleotides.
These chain-terminating bases are also labeled with dyes. Each base (A, T, C, G) will have a different color. This lets us see exactly which one is added.
Sanger sequencing uses a large amount of regular nucleotides, and a small amount of chain-terminating nucleotides. Since incorporation of the special bases is random, we'll have DNA fragments of various lengths with the special base at one end.
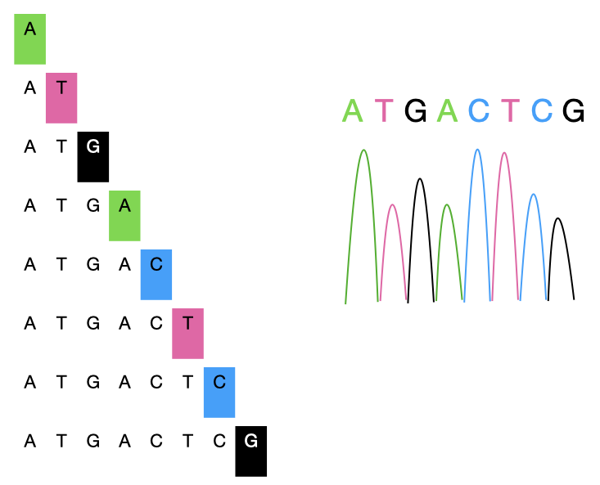
For example, let's say we're trying to sequence this piece of DNA: ATGACTCG.
If a chain-terminating 'A' base is incorporated to the DNA, the reaction will stop at the 'A' positions. So we have fragments: A and ATGA.
Similarly, if a special 'G' base is incorporated, the reaction will stop at the 'G' positions. So we have: ATG and ATGACTCG.
So we'll have a pool of DNA fragments, shown below on the left.
These fragments are then separated by size by a long glass capillary filled with gel. The speed at which these fragments move in the gel is dependent on their size. The longer the fragment, the slower it moves.
At the end of the capillary, there is a light sensor that detects the color emitted from the chain-terminating nucleotide. A computer program will then process the signals and then generate the sequence.
What is Next Generation Sequencing (NGS)?
"Next Generation Sequencing" (NGS) refers to a group of technologies that were introduced in the past decade. These newer technologies enabled rapid and high-throughput sequencing of DNA and RNA.
The most commonly used method is developed by Illumina. While there are other methods, that's the one I'll focus on here.
Illumina's technology is based on similar principles as Sanger sequencing. As in Sanger, dye-labeled nucleotides are added by DNA polymerase, and the colors are used to read the sequence.
But unlike Sanger Sequencing, NGS methods can sequence an entire genome's worth of DNA in one experiment. It can do this by running millions o PCRs at the same time, and looking at which base is added in each of those independent reactions.
Before you can do NGS, you have to prepare your sample for sequencing. First, all the DNA has to be cut into similar-sized pieces. Then you have to add in sequencing adaptors. These are like tiny handles that help hold onto the DNA during sequencing.
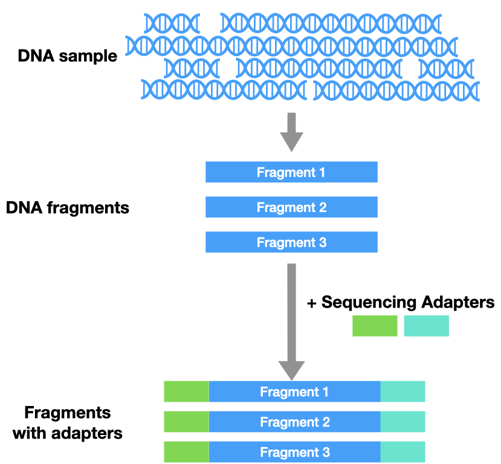
Instead of running a reaction in a tube, the DNA is loaded onto a flow cell. There are millions of tiny wells on the surface of a single flow cell. Each of these wells can capture a single piece of DNA from the sample, by grabbing onto an adaptor.
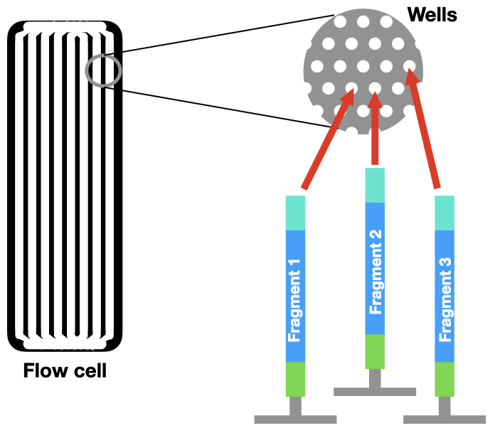
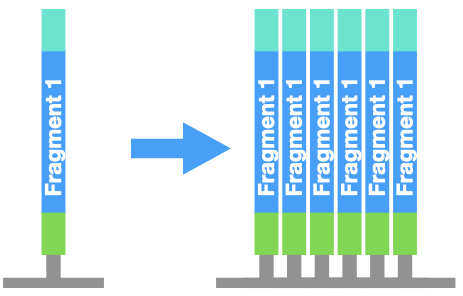
First, the DNA piece in each well is copied by PCR. This step is very important! It turns one molecule of DNA into a cluster of identical DNA pieces. This makes a denser "spot" that is easier for a computer to see.
The principle in Illumina sequencing is similar to Sanger sequencing. But instead of using those special chain-terminating bases, a dye-labeled regular nucleotide is used. Because of the dye, polymerase can only add one base at a time.
Once a base binds to the DNA, a picture of the flow cell will be taken. Since each nucleotide (A, T, C, or G) has a distinct color, we can find out which one it is. Then the dye is washed off and a second base binds to the DNA cluster. This cycle is repeated 100-200 times to get the complete sequence.
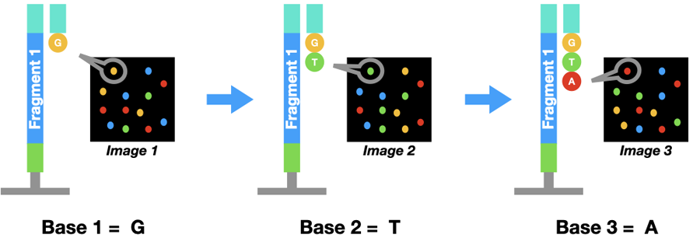
Since millions of pieces of DNA are sequenced at a time, you end up with a lot of data! Scientists generally use computer programs to help analyze all this information.
How is NGS compared with Sanger sequencing?
The basic principles behind NGS and Sanger sequencing are similar. Dye-labeled nucleotides are added to the growing strand of DNA, and each base is determined based on the color of the dye.
The biggest difference between the two is sequencing volume. Sanger sequencing can only sequence one fragment at a time. Because NGS uses flow cells that can bind millions of DNA pieces, NGS can read all these sequences at the same time. This high-throughput feature makes it very cost-effective when sequencing a large amount of DNA.
The other main difference is sequencing length. A single sequence generated by NGS methods will be 100-200 bases long. In contract, Sanger sequences will be 700-1000 bases long.
Sanger sequencing is a good choice when sequencing a short region in a small number of samples. But if you need to sequence an entire genome (or a lot of samples), NGS may be the more cost-effective choice.

Author: Allison Zhang
When this answer was published in 2019, Allison was a postdoctoral fellow in the Department of Genetics, studying the Exposome (how environmental exposures affect your health) in Mike Snyder’s laboratory. She wrote this answer while participating in the Stanford at The Tech program.
 Skip Navigation
Skip Navigation
