
Y am I seeing Y DNA in my genetic test results?
February 25, 2022

- Related Topics:
- Genotyping,
- Ancestry tests,
- Consumer genetic testing,
- Trisomy/Aneuploidy
A curious adult from Arizona asks:
“I am a female, yet I notice my 23andMe test results report has Y DNA. How common is this? Does it mean I am xxy or xy or that some of a y chromosome is attached elsewhere in my genome? How can I tell?”
We get this question a lot at Ask-a-Geneticist! In many cases, this is simply due to some confusion in how results are displayed in the raw data browser.
The type of test 23andMe uses isn’t particularly good at detecting differences in chromosome number. And they’re pretty up front about this, listing XXY (Klinefelter’s syndrome) and other chromosome differences as conditions not included in 23andMe reports. Their algorithm just isn’t designed to analyze unexpected chromosome differences.
While it might be possible to figure out you’re XXY from such a test, you’d definitely want to take a different type of medical-grade genetic test to confirm.
One chip fits all
Normally, the test starts with a saliva sample that contains your genomic DNA. Your sample contains A LOT of information about you. Your genome is approximately 6.2 billion base pairs (6,200,000,000 letters!) in length. If you read a base of your genome every second, it would take you almost 100 years to finish!
Because of this, most ancestry-type tests use a method called genotyping. Instead of looking at every single one of the 6 billion letters in your DNA sequence, they only look at a million or so spots.
They pick these specific spots because they are known to vary between different people. For example, one person might have an "A" at a specific position, while another person might have a "T" at a specific position. This is called a single nucleotide polymorphism (or SNP, pronounced "snip").
Testing these millions of SNPs one by one would take a while. Instead companies test all of them at once using a genotyping chip. Each chip contains millions of tiny “probes”, which light up if they find matching DNA. Each chip will have one probe for every variant of SNP being tested – both the ones you have and the ones you don’t.
Since only a single genotyping chip is used for everyone who takes the test, the chip must contain probes for both X and Y chromosomal DNA. That means that your sample will be tested for Y-DNA no matter what chromosomes you have.

The raw data browser
Shown below is an image of what your raw data dashboard might look like If you used a testing service like 23andMe. The confusing part is that it looks the same for everyone who takes a 23andMe test. No matter what chromosomes you actually have, the graphic will show a single X chromosome and a single Y chromosome.
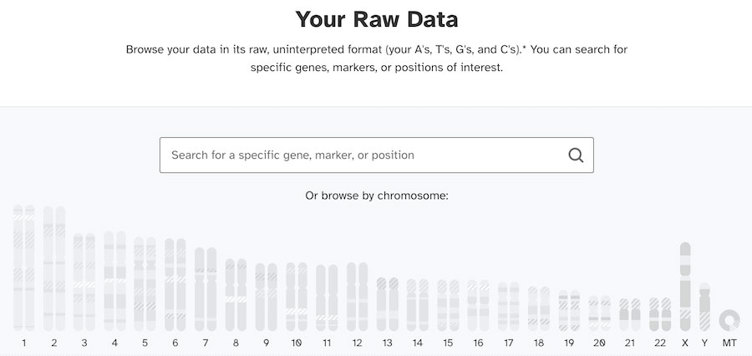

As we mentioned earlier, the genotyping chip tests for Y chromosomal DNA in everyone. That means that everyone’s report will include some parts of the Y chromosome.
But when a person without a Y chromosome clicks on the image to see the results, here’s what that looks like:
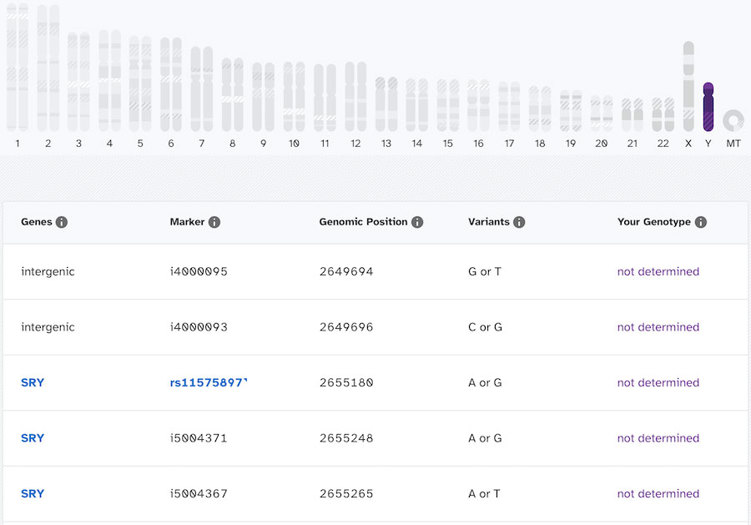

Looking at these, even though you can see all the markers and genes on the Y chromosomes, the actual genotype call is “not determined” for all of these. “Not Determined” means that a confident genotype couldn’t be assigned at that position. In other words, they couldn’t say for sure whether the variant was there or not.
In contrast, here’s what it looks like for someone who does have a Y chromosome:
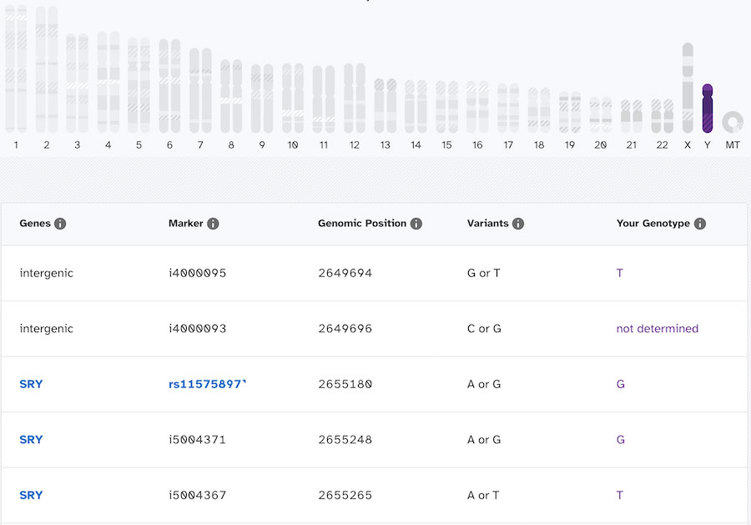

You can see that this person has actual genotype calls for most of the markers on the Y chromosome. There are a few “not determined” spots, but most of the spots have an A, T, G, or C. So you might be able to see now how there is a very large difference in what it looks like when a Y chromosome is present compared to when one is absent.
Y you might see a false-positive
Now let's get back to the results of your test. It’s actually very common for someone without a Y chromosome to see some Y-DNA on their raw test results.
If you were to keep looking through all of the Y-chromosomal data in the first test report from above, you might find a couple of spots with genotype calls. But remember that thousands of genotypes are normally identified in a report when a Y-chromosome is present.
In these cases, the small number of variants identified on the Y chromosome are probably false positives. That is what we call it when a test detects something that’s not really there.
We should be a bit skeptical of at home genetic testing results. 23andMe says that its accuracy rate is greater than 99%. Put another way, 1% of results can be errors. So out of the 650,000 positions they report per person, you would expect about 6,500 of them to be wrong!
When we compare this rate to the rate of having XXY sex chromosomes (0.1-0.2%), we can see that just by this comparison alone, it is far more likely that the Y-chromosomal DNA detected in your raw genotyping results are false-positives rather than a true case of Y-chromosomal DNA actually being in your sample.
To understand how the test can “find” some Y-DNA in your sample that’s not actually there, let's think back to the genotyping chip that I mentioned earlier. Remember how I said the chip is covered in tiny probes that look for DNA matches? It turns out that your X-chromosomes have parts that are very similar to Y-chromosomal DNA. This means that probes “looking for” Y-DNA might get confused by this, and incorrectly pair with X-chromosomal DNA.
And this false positive problem isn’t just limited to the X and Y chromosomes. You can have false positives (or false negatives!) in other parts of your test results as well. This is actually very common in at-home genetic tests.
Detecting XXY?
But let’s ignore the false positives and the potentially confusing graphics for a second. Could you detect whether you’re XXY through a test like this?
Unfortunately, probably not. Genotyping tests aren’t great at detecting chromosome number differences like this. As 23andMe itself warns, their methods just aren’t designed to detect conditions like Turner syndrome (45,X) and Klinefelter syndrome (47,XXY).
Having said that, it’s possible that someone with XXY chromosomes might be able to see a hint of that in their results. In this case, you’d be looking for two things:
- Genotype calls all along the Y chromosome (like in the XY individual above)
- Pairs of genotype calls all along the X chromosome (like in the XX individual below)
Here’s what the X chromosome results look like for an individual who has two X chromosomes:
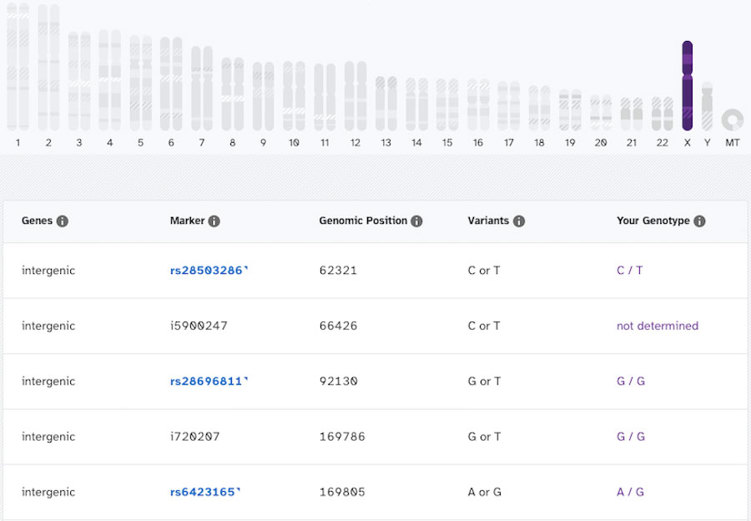

Even if you do see a result like that, you’d want to get a medical diagnostic test to confirm. In particular, a karyotype test would be the best way to detect chromosome differences. A doctor or genetic counselor could help with this.
Hopefully this helps explain why there are a lot of problems with looking too closely or “over-analyzing” the results of at-home genetic testing. These tests can sometimes be misleading!
Read More:
- How large is the human genome?
- Scientific American: How Accurate Are Online DNA Tests?
- 23andMe: How 23andMe Uses Your Self-Reported Sex and Gender
- 23andMe: What Does "Not Determined" Mean?
- New York Times Opinion: Why You Should Be Careful About 23andMe’s Health Test

Author: Erin Brooks
When this answer was published in 2022, Erin was a research assistant in the Departments of Genetics and Hematology. She studies the role of the gut microbiome in health and disease in Dr. Ami Bhatt’s laboratory. She wrote this while participating in the Stanford at The Tech program.
 Skip Navigation
Skip Navigation
