
How can you tell two different DNA samples apart from each other?
May 23, 2012

- Related Topics:
- DNA sequencing,
- Genetic testing,
- Bioinformatics
A middle school student from California asks:
"How can you tell two different DNA samples apart from each other?"
You can’t just look at two DNA samples and tell if they’re different or not. They’ll both look like a ball of snot whether they come from people, dogs or daisies.
And a microscope won’t help either. All DNA will look alike even with the most powerful ones.
But there are many chemical ways to tell two DNA samples apart. I’ll focus on just two of them—RFLP analysis and DNA sequencing. Either method would quickly tell my DNA from yours. Or yours from a dinosaur’s!
To understand how each works, we need to take a step back and talk about what DNA is. And how your DNA is different from mine.
Lots of Ways to be Different
DNA is made up of four different letters. The four letters are A, T, G, and C. Humans have 6 billion of these all laid out in a certain order.
All humans share about 99.5% of the same DNA. But that leaves plenty of differences between your and my DNA.
Sometimes there is just a single letter difference. You might have a G at a certain spot while I have an A.
Your DNA: ATGGATGC
My DNA: ATGAATGC
Scientists call these differences SNPs (pronounced “snips”). On average you and I have one of these every thousand letters or so.
There can also be larger changes between two DNA samples. You can have an insertion, which means you have extra letters.
Your DNA: ATGGTTGAATGC
My DNA: ATGGATGC
You can also have a deletion, which means you are missing letters that I have.
Your DNA: ATGATGC
My DNA: ATGTTTGGGAAATTTGGATGC
As you can see there are many ways your DNA could be different from mine. And there are also many ways to tell my DNA apart from yours.
I will be describing two common ways scientists can use to tell whether or not DNA samples are different. The main idea of each of these techniques is to recognize the differences in the letter sequences between two DNA samples.
RFLP Analysis
Restriction Fragment Length Polymorphism (RFLP) is a long name for a pretty simple technique. Basically scientists use certain enzymes from bacteria to cut up two samples of DNA.
These enzymes cut DNA at very specific DNA sequences. Scientists cut different DNA samples with the enzyme and look at the pattern of cuts. If the pattern of cut DNA looks different, then the two samples were different.
Well, maybe that didn’t sound so simple. Let me give a couple of examples to show you what I mean.
Example 1: Different DNA Sequences Will Have a Different Number of DNA Pieces
Certain bacteria make an enzyme called EcoR1. This enzyme likes to cut DNA at GAATTC.
Let’s say that your DNA looks like this is a certain section:
GATCTCGAATTCGGTAACCTTAACCGTGTGAACTGGAATTCATTATTATTTTAGCGC
And mine looks like this:
GATCTCGAATTCGGTAACCTTAACCGTGTGAACTGGATTTCATTATTATTTTAGCGC
Notice that there is one letter different. Notice also that it changes a GAATTC to a GATTTC. In other words, you have two places where EcoRI likes to cut, while I only have one.
When each of our DNAs is treated with EcoR1, we’ll end up with a different pattern of cut DNA. You’ll have 3 fragments that are 6, 35 and 16 bases long. I’ll have just two fragments that are 6 and 51 bases long.
Example 2: Different DNA Sequences Will Have Different Sized Fragments
This technique can also be used to see missing or extra DNA.
Imagine your DNA looks like this:
GATCTCGAATTCGGTA----------------CGTGTGAACTGGAATTCATTATTATTTTAGCGC
The dashed lines are missing DNA. And my DNA looks like this:
GATCTCGAATTCGGTAACCTTAACCGTGTGAACTGGAATTCATTATTATTTTAGCGC
Now when we cut with EcoRI, I get 3 fragments that are 6, 35, and 16 letters long. You get three fragments too. But yours are 6, 29, and 16 long.
In both cases we were able to see the differences on paper. But how can scientists actually see these differences in real life? By taking advantage of the different physical properties of big and little pieces of DNA.
How To See The Different Fragment Patterns From RFLP Analysis
There are many ways to separate DNA based on size. One of the most common techniques is called gel electrophoresis.
In gel electrophoresis, scientists load the DNAs into special wells in a gel. Then, with the help of electricity, they separate the DNAs by size. Here’s what the gel looks like:
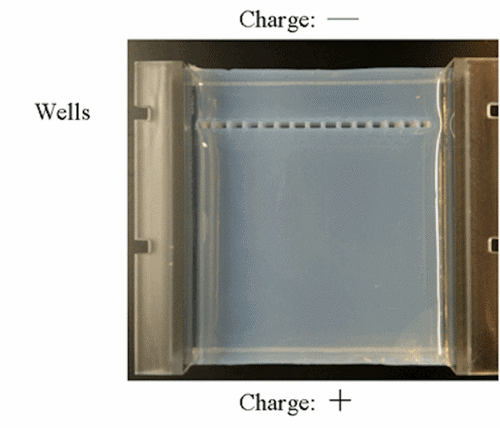

Notice the small rectangles in the gel. Those are the wells where DNA is loaded.
Scientists then use electricity to force the DNA through the gel. Since DNA has a negative charge, it runs away from the negative side and towards the positive side.
What happens is that the smaller pieces move through the gel faster than the big ones. In the end, you get a certain pattern based on the sizes of the DNA fragments in the gel.
To understand why bigger pieces move more slowly, we need to look at a microscopic view of the agarose. Here it is:
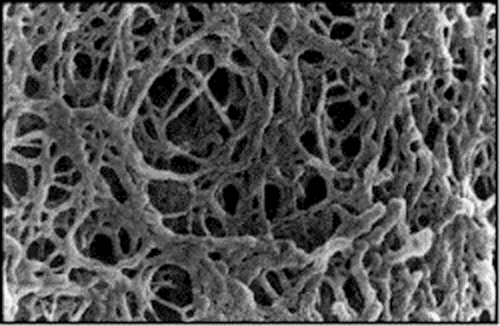

As you can see from the picture, the gel has lots of small holes in it. So when scientists put their cut up fragments into a gel, smaller pieces move faster and further than larger pieces. It is harder for larger pieces to squeeze through the small holes in a gel.
Remember our examples from the previous section? Here is what they would look like on agarose gels:
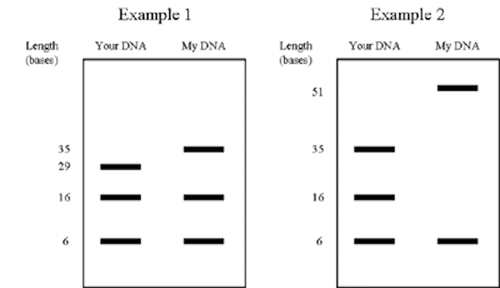

The bigger pieces are higher up in the gel compared to the smaller pieces. As you can see, you can definitely tell them apart!
RFLP analysis is a pretty useful technique for telling us that two DNAs are different. But it can’t always tell us how they’re different. This is where the other technique, sequencing, comes in handy.
Sequencing
Scientists can also tell if two DNA samples are different by sequencing. Sequencing will definitely tell you if two DNAs are different AND how they are different. It will tell you the exact letters in a DNA sample.
I won’t go into all the details of sequencing here. But see our previous answer for more details if you’re interested.
So when you sequence DNA, you’ll get the letters of the DNA directly.
Your DNA: ATGCATGC
My DNA: TTATGCATGC
In the example above you learn that your DNA and my DNA are different. You also learn that my DNA has two extra Ts compared to yours. RFLP analysis would not tell you that.
Analyzing Sequencing Data
As you can imagine, sequencing can give you a lot of information. If you had really long DNAs, then you could end up with pages and pages of DNA letters.
These are a real pain to go over by eye. Imagine comparing all 6 billion of your and my letters one letter at a time. I can see scientists leaving the field in droves…
Luckily scientists have come up with computer tools that can help us with this tedious analysis. The National Institutes of Health has online tools that can help you see if and how two long DNAs are alike or different.
There is BLAST: Basic Local Alignment Search Tool. In BLAST, you can copy and paste two DNAs into the BLAST form. You hit ENTER and BLAST will then do some calculations. It will show you the parts of the DNAs that are exactly alike, similar, and different.
BLAST can also tell you how alike your DNA is to DNA sequences from other different living things (daisies, dogs, bacteria). And it can help you figure out where an unidentified piece of DNA came from.
Summary
There are many other ways you can tell two different DNAs apart. One way is to cut up the DNAs and see if they have the same fragment patterns. Another way is to sequence the DNAs. The key differences you will be looking for are differences in the DNA sequences.
Read More:
- NIH Basic Local Alignment Search Tool (BLAST): https://blast.ncbi.nlm.nih.gov/BlastAlign.cgi
- Science Buddies: Gel electrophoresis
- Arstechnica: Brief guide to DNA sequencing
- Jove: Step by step video on how to run a gel (gel electrophoresis)

Author: Mei-Hsin Cheng
When this answer was published in 2012, Mei-Hsin was a Ph.D. candidate in the Department of Genetics, studying epigenetic regulation in drosophila in Joseph Lipsick’s laboratory. Mei-Hsin wrote this answer while participating in the Stanford at The Tech program.
 Skip Navigation
Skip Navigation
