
What makes a gene a gene?
May 8, 2013

- Related Topics:
- DNA basics,
- Genes to proteins,
- Bioinformatics,
- Noncoding DNA
An undergraduate from California asks:
“What makes a gene, a gene? I know it needs to have nucleotide sequences such as TATAAA that encode for proteins and a gene needs to also have a function but is this it?”
That is a very good question. You might think that scientists would have a clear idea by now about what a gene is. After all, the human genome was sequenced over ten years ago, and we have learned a great deal more about biology and genetics since then.
But it seems like the more we learn, the less sure we become of what a gene actually is. The traits you’ve listed are a great start for what we would have defined as a gene ten or twenty years ago.
Back then, we would have said a gene has to have a certain DNA sequence, called a promoter, at the beginning that identifies it as a gene. Some promoters do have the TATA box you mention, but many do not. Still, there are certain bits of DNA that we can recognize as a promoter.
A gene was also thought to have the instructions for a functional protein. A protein is simply something that does a specific job in a cell. Hemoglobin, the protein that carries oxygen in our blood, is an example of one of these.
However, in the last decade or so we’ve realized that this is way too narrow a definition of what a gene is. There seems to be lots of DNA that is important for getting jobs done in the cell that do not do so by making a protein.
What this boils down to is that after all these years, scientists still aren’t sure how to define a gene! Let’s dig a bit deeper to see what is going on with the DNA in our cells.
The Central Dogma, or How DNA is like Books in a Library
Your DNA consists of four different types of bases: adenine, thymine, guanine, and cytosine (often abbreviated to A, T, G and C). These four bases are strung into a sequence that is over 6 billion bases long to form your DNA.
We can think of a traditional gene as a recipe from a cookbook in a library. But instead of looking for recipes for a dish like spaghetti or cheesecake, we are looking for recipes for a protein.
Your cells need to make proteins in order to function and stay alive. When a cell needs a certain protein to be made, it first copies some of the instructions contained in the DNA into another molecule, RNA. This process is called transcription. This would be like going to a library and photocopying a recipe that you want to make from one of the cookbooks.
When a cell gets a part of its DNA transcribed, the “copy” is called mRNA. For the most part, the mRNA and DNA contain the same message, just like an actual photocopy of a recipe.
The mRNA then gets read, or translated, into the actual protein – the functional product that the cell needs to carry out its jobs. This is like going home with your photocopied recipe and making the dish. Your resulting dish would be like the protein.
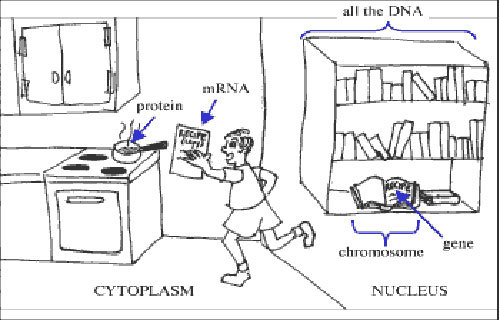
The process of going from DNA to RNA to protein is known as the Central Dogma of biology.
Genes were originally thought to be the parts of DNA that code for a protein, or the “recipes” contained in cookbooks in a library. This is because scientists believed that proteins did almost all of the work for cells. As such, when trying to identify genes, they first tried to find protein-coding regions of DNA.
Searching for Genes
One thing scientists have done to find potential genes is to look for possible promoters in DNA. As I mentioned in the introduction, a promoter is a specific DNA sequence that often appears at the start of a gene. It is important for determining whether the gene gets turned “on” or “off”.

In this sense, a promoter is sort of like the title of a recipe. Scientists notice that many genes share similar promoter sequences, so they’ve searched for these sequences in DNA to identify possible new genes. The TATAAA you mentioned is one of these common sequences.
Another way that scientists have looked for genes is to find long open reading frames (or ORFs) in DNA. An ORF is a stretch of DNA that could possibly code for a protein. There are specific sequences in your DNA that signal where the protein-coding region starts (ATG) and where it ends (TAG, TGA, TAA). If these start and end signals are a certain distance away from each other, that suggests the presence of a gene.
Since there are rules like this for gene identification, you might wonder why it would be difficult to determine what a gene is. In simple organisms like bacteria, these rules for finding genes hold pretty well. This is because bacterial promoters are not very complicated, and there are no break-points within genes.
In humans, however, identifying genes gets a little more messy. This is partly because promoters of genes in humans tend to be more diverse, both in their sequence and also in how far away they are from a gene. There are also sequences within genes in humans that don’t end up getting translated into protein, which are called introns.
So, instead of a nicely composed recipe to read off of, as you have in an organism like bacteria, there might be a lot of notes written in between the steps, or whole parts that are crossed out. This all makes it more difficult to find genes in the human genome.
Genes That Don’t Code for Proteins?
Scientists originally considered a gene to be something that codes for a protein, as these were considered to be the functional molecules in a cell. However, some pretty recent research has led us to believe otherwise.
A large research endeavor, known as the ENCODE consortium, recently found that about 80% of our DNA gets transcribed into RNA. However, only 2-3% ends up getting translated into protein. This means that there is a lot of non-coding RNA floating around in our cells.
Going back to our library example, this is like making photocopies of pages in novels. You can read them, but they won’t give you instructions for making a dish.
How much of this non-coding RNA has a function? This is an active area of research for many labs right now. We do know that some of this non-translated RNA is very important. For instance, some types of RNA play a role in controlling how much protein gets made by a gene. One way they can do this is by binding to the RNA that is copied from a gene and either helping or (more often) hindering that RNA from being translated into protein.
It’s clear that at least some non-coding RNA is performing important tasks for the cell, but should the DNA coding for it still be considered a gene? And how do we even determine whether a particular non-coding RNA has a function or not? Although the concept of a gene has been around for over 100 years, the details of defining what it is are still being worked out!
Read More:

Author: Arielle Yablonovitch
When this answer was published in 2013, Arielle was a Ph.D. candidate in the Department of Biophysics, studying RNA editing in drosophila in Billy Li’s laboratory. She wrote this answer while participating in the Stanford at The Tech program.
 Skip Navigation
Skip Navigation
